nCoV-2019 novel coronavirus RAMPART runtime instructions
Nanopore | bioinformatics
| Document: | ARTIC-nCoV-RAMPART-v1.0.0 |
| Creation Date: | 2020-01-23 |
| Author: | Andrew Rambaut |
| Licence: | Creative Commons Attribution 4.0 International License |
- This document is part of the nCoV-2019 Nanopore sequencing protocol package:
- http://artic.network/ncov-2019
Related documents:
- nCoV-2019 Nanopore bioinformatics environment setup:
- http://artic.network/ncov-2019/ncov2019-it-setup.html
Preparation
Set up the computing environment as described here in this document: ARTIC-nCoV-ITSetup. This this will install RAMPART as part of the conda environment.
Activate the ARTIC environment:
All steps in this tutorial should be performed in the artic-ebov conda environment:
conda activate artic-ncov2019
You can check that RAMPART is installed:
rampart --version
The current stable release is v1.0.3.
Preparing for a sequencing run:
Our recommended procedure for a new sequencing run is to create a directory for that run. We suggest using the same name as the Experiment name you provide in MinKNOW:
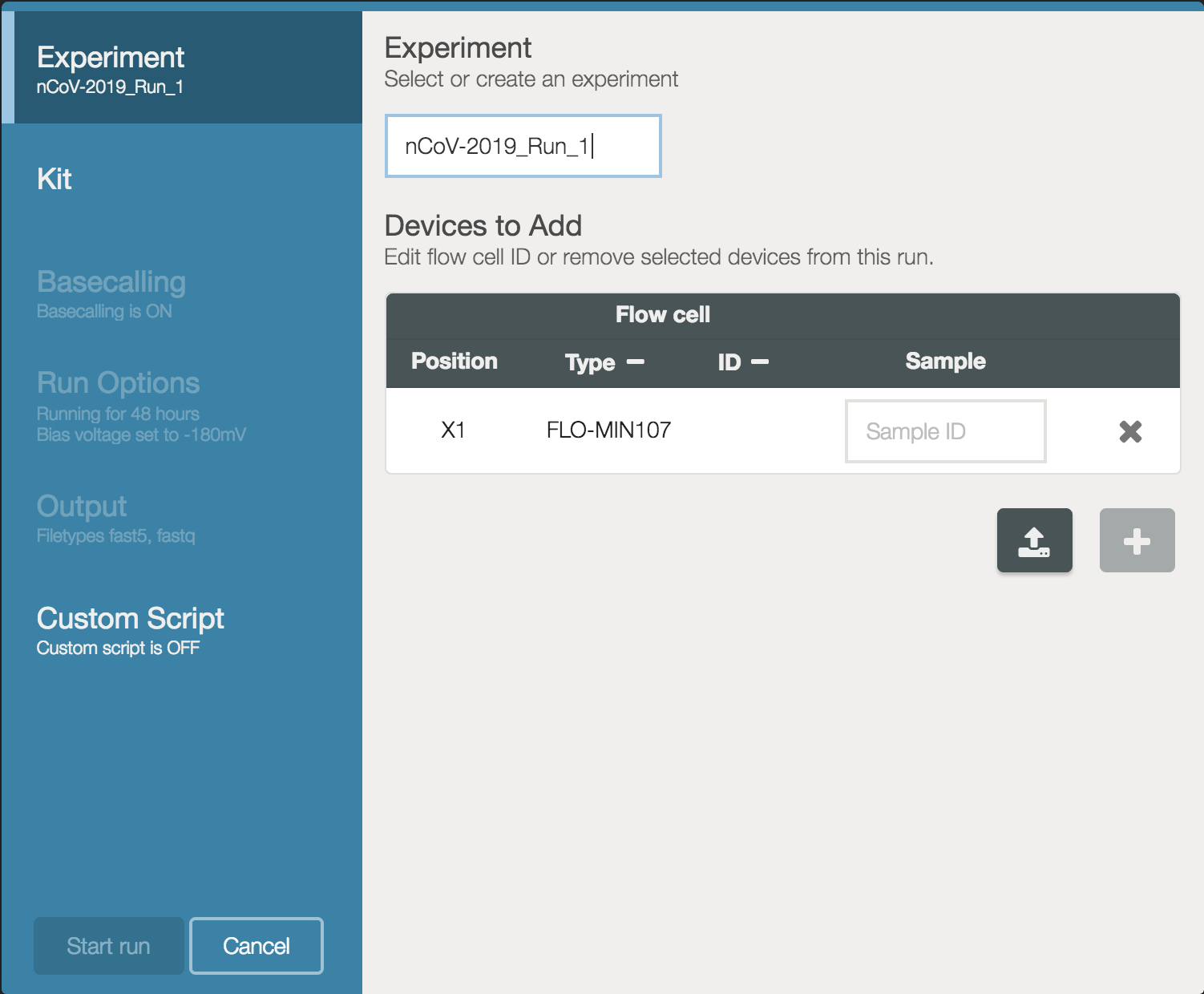
The experiment name will be referred to here as <run_name>.
mkdir nCoV-2019_Run_1
cd nCoV-2019_Run_1
By default MinKNOW writes 4000 reads to each FASTQ file it creates. If you want finer scale monitoring in RAMPART we suggest altering the settings to write 1000 reads per FASTQ file.
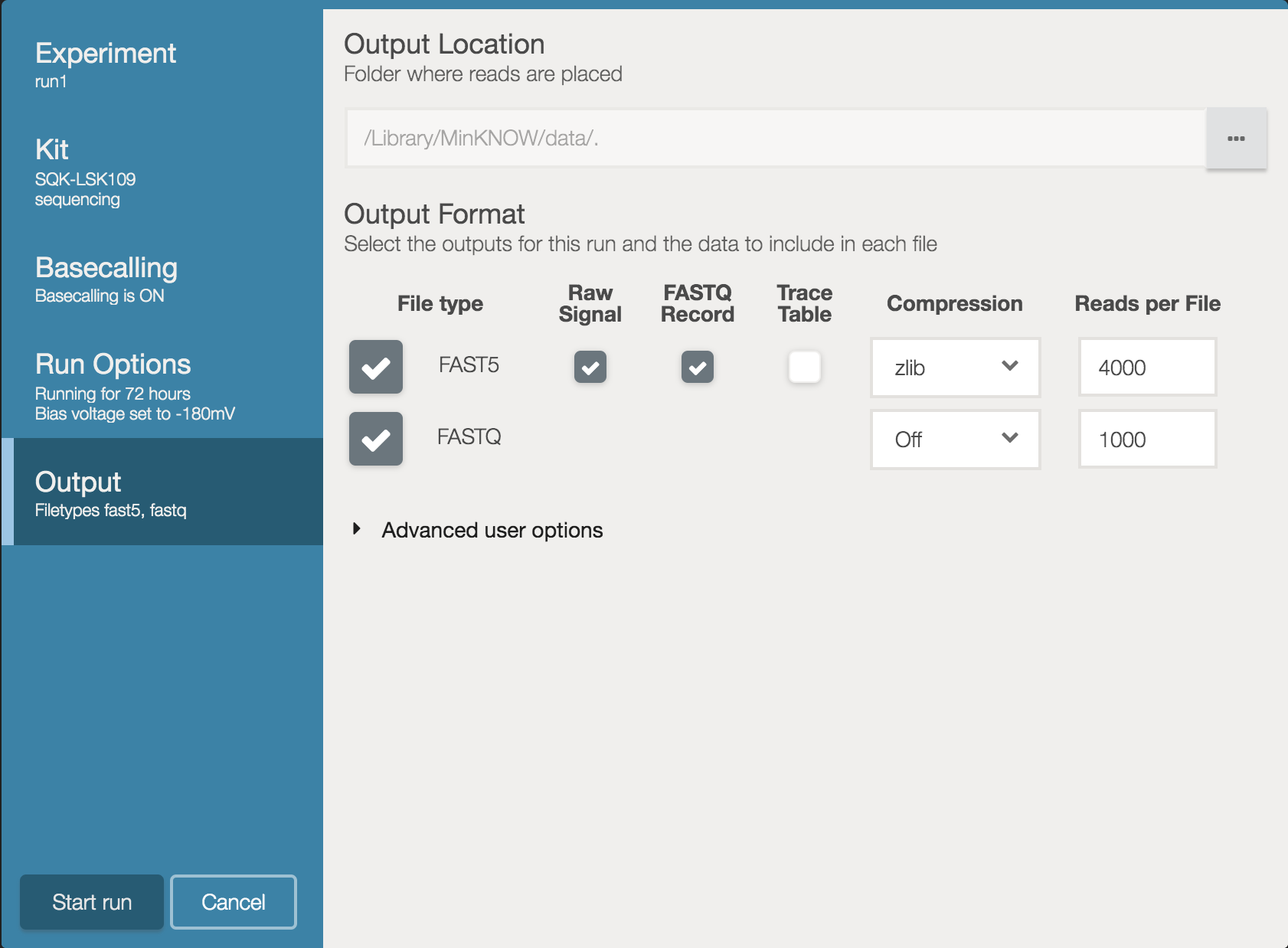
IMPORTANT – At the moment RAMPART is not compatible with MinKNOW’s built-in barcode de-multiplexing. This must be turned off.
Also take note of where the data is going to be written (the Output Location at the top of this window) as it can vary depending on your operating system. You can also change this to a different location (such as an external SSD).
This location will be referred to as <minknow_data_path>.
If you are using ONT native barcoding to multiplex multiple samples then you can optionally create a barcodes.csv file to provide the mapping of barcode to sample name. The barcodes.csv has one line per barcode and you should only specify barcodes that are actually present in your library. The file content looks like this:
barcode,sample
NB01,"Sample_1"
NB02,"Sample_2"
NB03,"Sample_3"
NB04,"Sample_4"
Running RAMPART
Start the sequencing run in MinKNOW and wait for the first reads to be basecalled.
Now start RAMPART:
rampart --protocol ../artic-ncov2019/rampart --basecalledPath <minknow_data_path>/<run_name>/fastq_pass
Finally start a browser (we recommend Chrome but most modern browsers should work) and point it to http://localhost:3000/ in the URL box.
The RAMPART header screen will appear but you may have to wait for sufficient data to be sequenced and basecalled before any results appear on the screen. RAMPART will only start processing FASTQ files when the second one appears so you will need to wait for 2000 reads to be base called (or 8000 if you left the reads per file at its default).
For more details about RAMPART visit: https://artic-network.github.io/rampart/.
RAMPART is open source and is on GitHub: https://github.com/artic-network/rampart/.