Setting up and using EPI2ME for MPXV analysis pipelines
ARTIC pipelines | bioinformatics
| Document: | ARTIC-MPXV-EPI2ME-Setup-v1.0 |
| Creation Date: | 2024-08-22 |
| Author: | Lauren Lansdowne, Andrew Rambaut |
| Licence: | Creative Commons Attribution 4.0 International License |
Requirements:
- The ability to install software on your desktop or laptop. This includes some extra packages like Java and Docker that may require administrator level privileges.
- Internet access when first downloading each pipeline, and for the first time running it. After that, you should be able to run it offline.
Installing the EPI2ME software
EPI2ME is a standard desktop software package available for Windows, Macintosh and Linux operating systems and is used to run ‘NextFlow’ pipelines in a simple easy-to-use environment without interacting with a command-line. It can be obtained from the EPI2ME website — https://labs.epi2me.io — with downloads for each operating system available from the download page:
Once you have downloaded the appropriate installer for your operating system, install it in the usual way.
Initial setup of EPI2ME
When you run EPI2ME it will ask if you want to sign-up or sign-in to the Cloud-based service — you can just click the Continue without signing in button.
The first time you run EPI2ME it may need to install some additional software. It will guide you through this process.
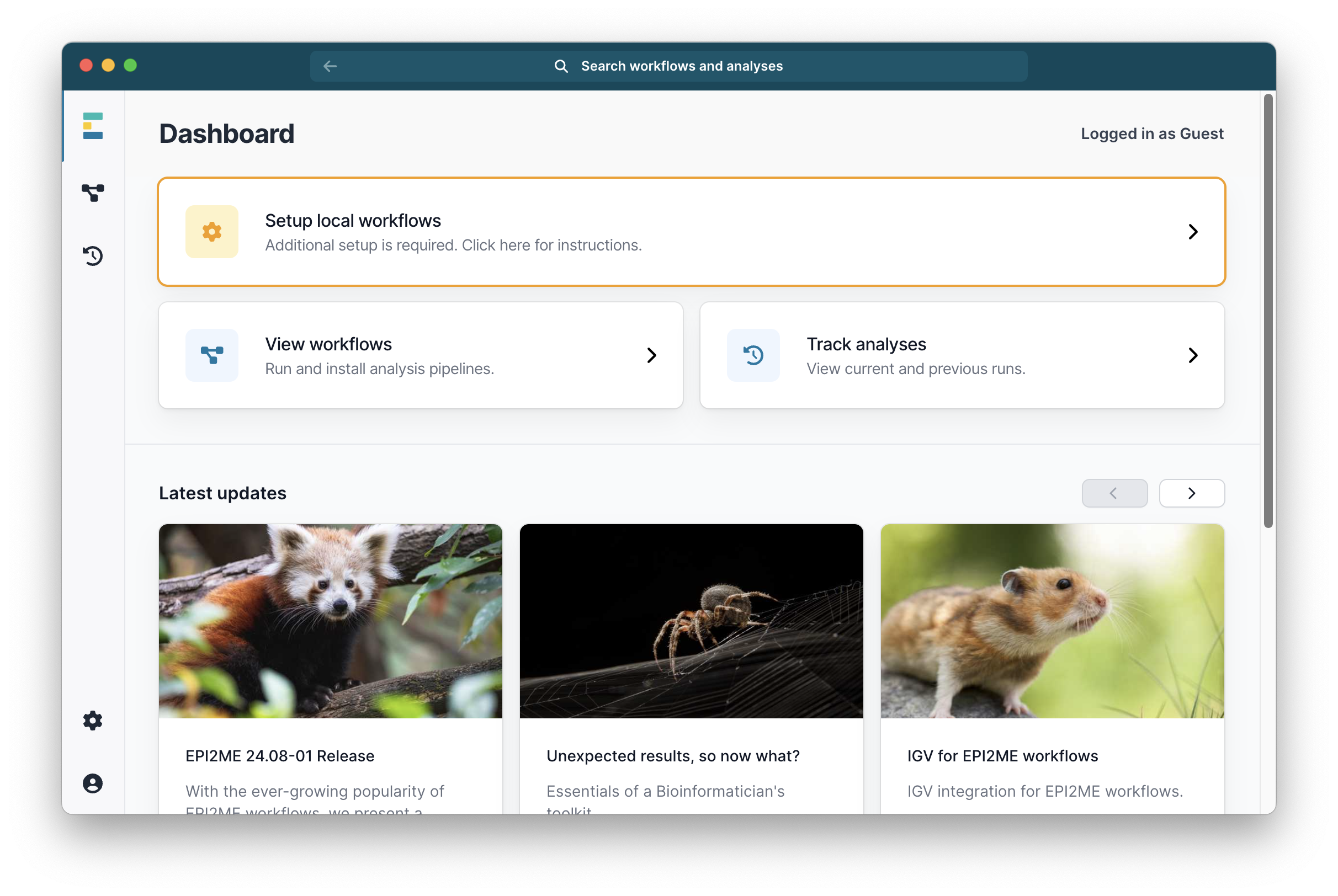
If you see a panel that says Additional setup is required then select it:
You may need to install the NextFlow software and Java - in which case click the Setup button:
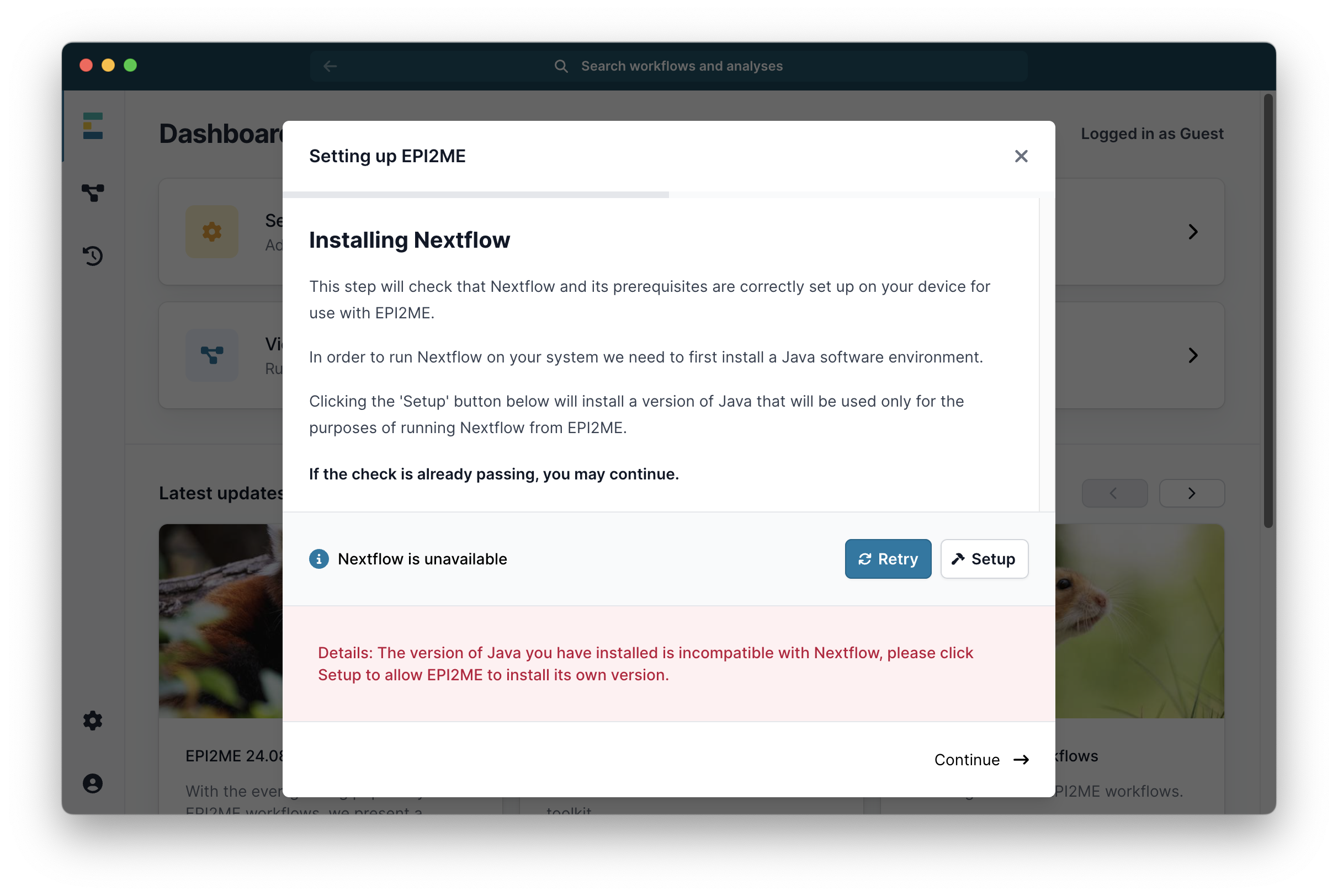
Then click Continue ->. Unless it has already been installed it will next ask you to install the Docker software. Click on the link to Docker Desktop Install and follow the instructions there to install Docker:
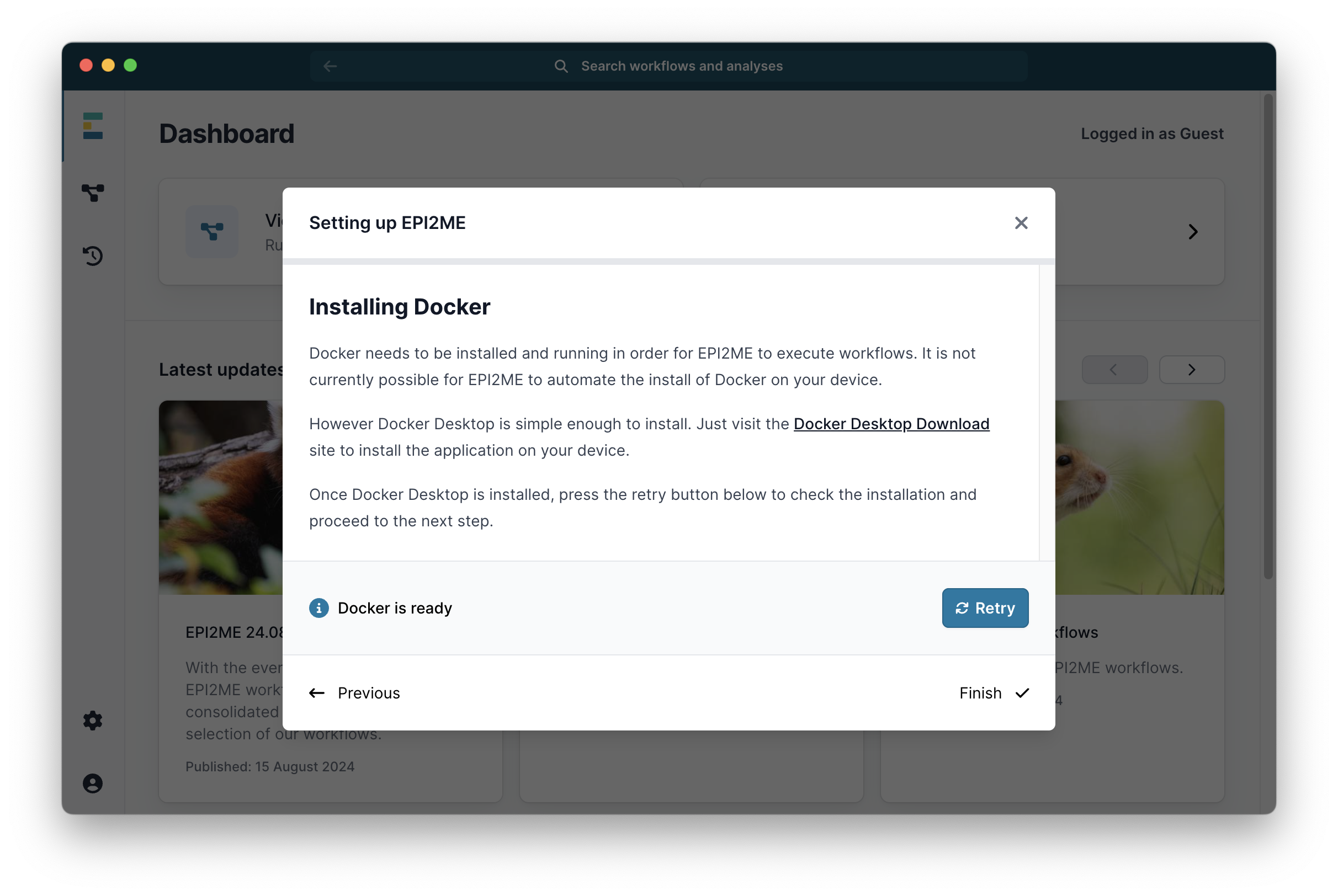
When it says Docker is ready, click Finish. The EPI2ME window will now look like this:
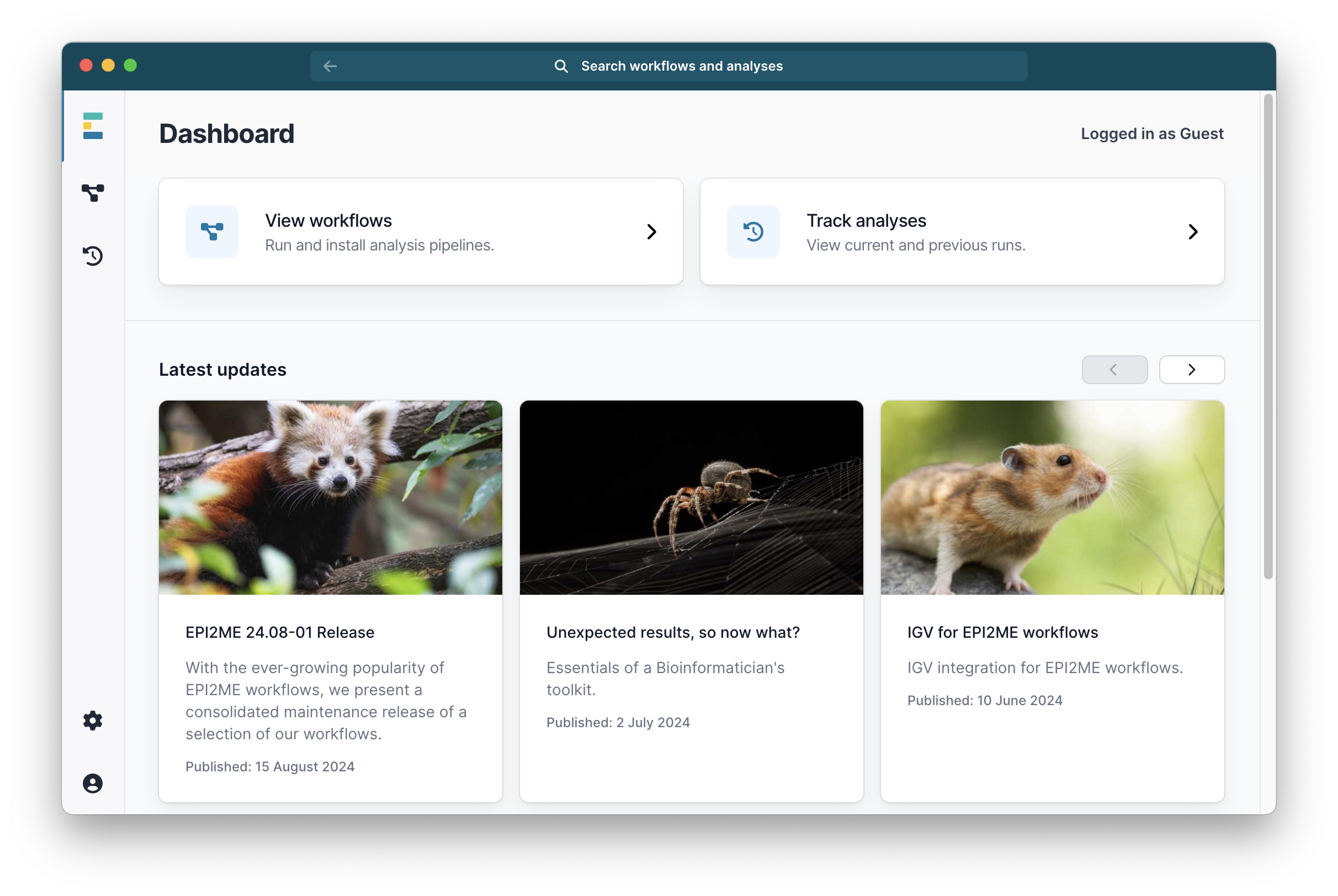
You are now ready to install an analysis pipeline in EPI2ME.
Using an ARTIC analysis pipeline in EPI2ME
Import the workflow
Open EPI2ME. On the main dashboard select “View workflows”.
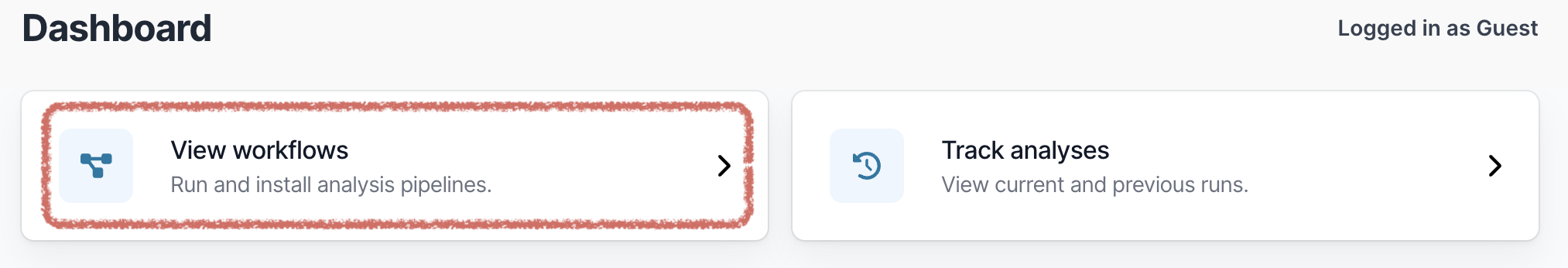
Then select “Import workflow”.
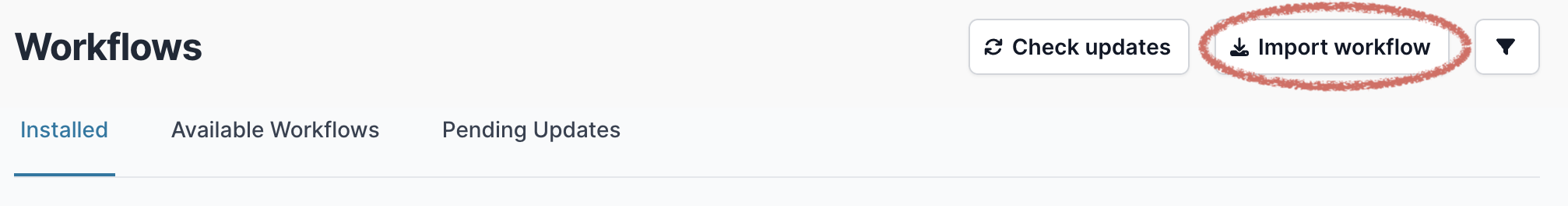
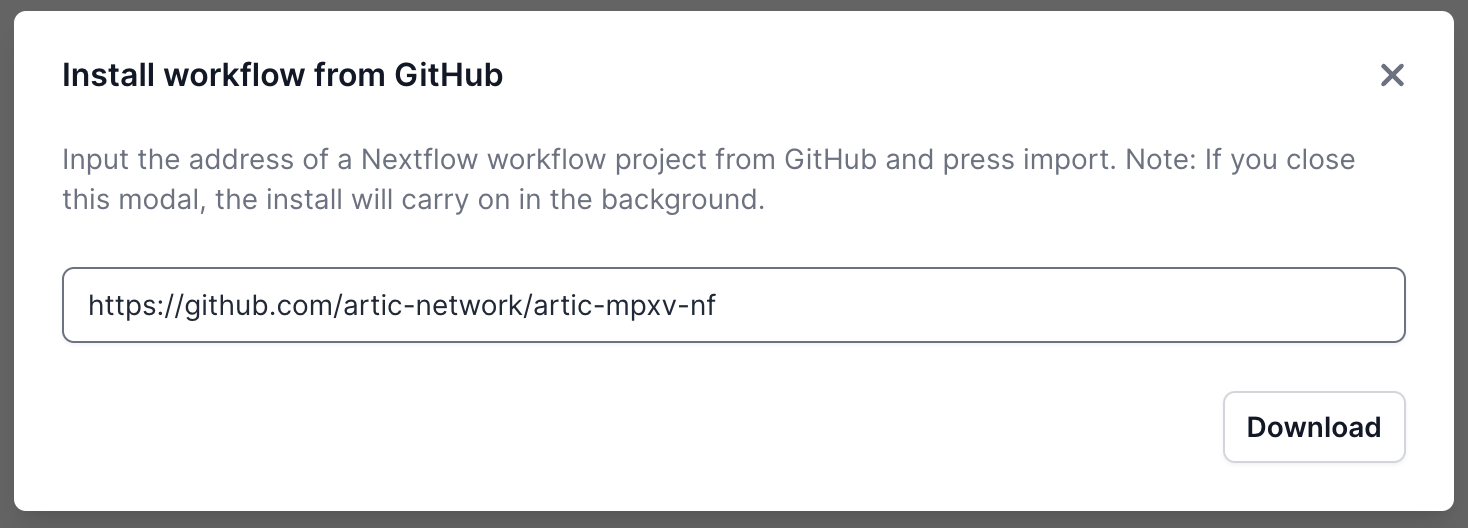
A pop-up window will appear where you can enter the GitHub URL. Enter the URL and click “Download” — the exact URL will depend on the pipeline you wish to install. For example, to install the ARTIC bioinformatics pipeline for nanopore sequence data use https://github.com/artic-network/artic-mpxv-nf (details about running this pipeline can be found here):
Once it has downloaded, it will be ready in the Installed tab:
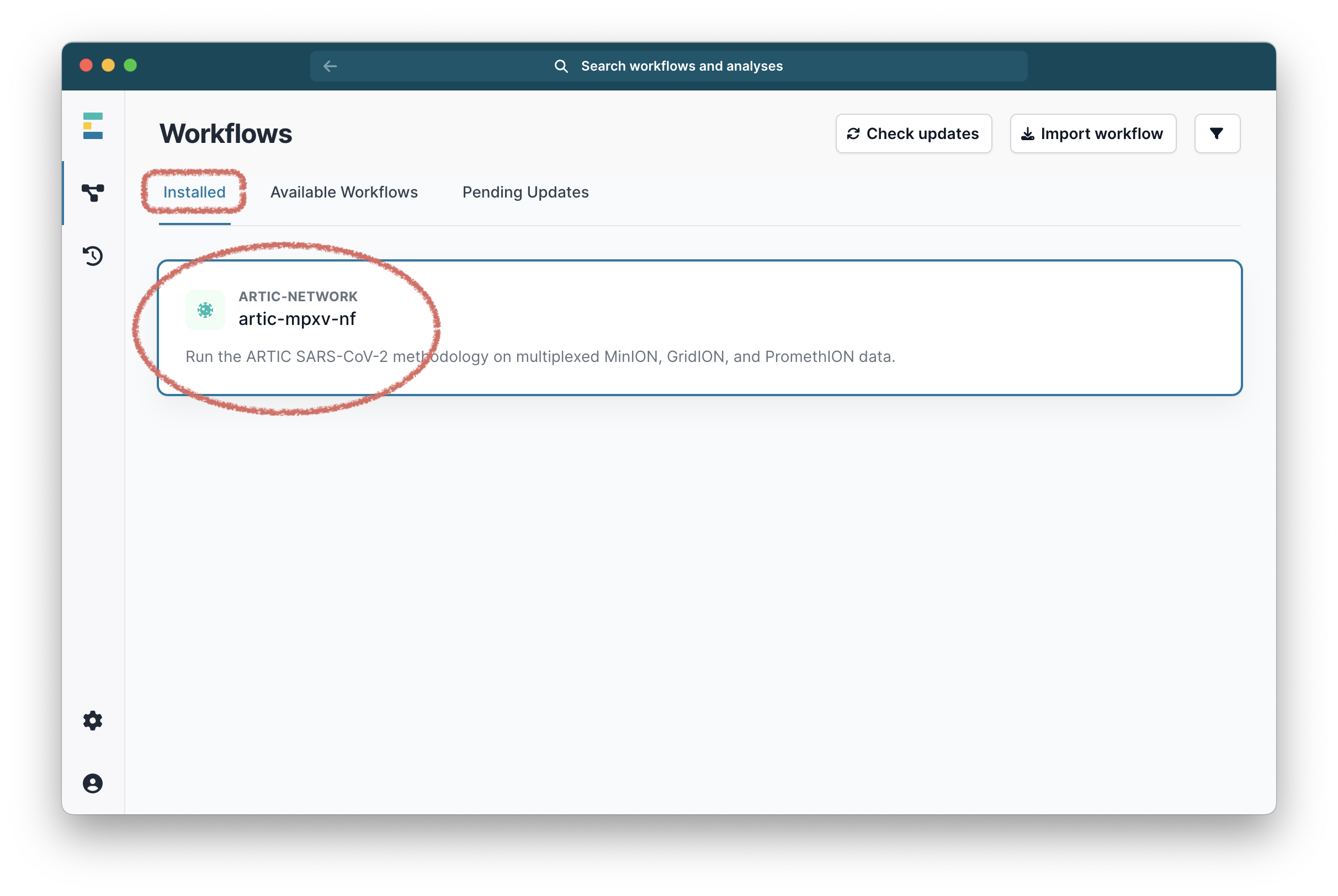
Select it and you will be taken to a landing page for this workflow.
Software credits
This resource depends on the EPI2ME desktop software provided by Oxford Nanopore Technologies. The pipeline is written in Nextflow and relies on Docker containers.
Running a pipeline
For instructions on running individual ARTIC pipelines in EPI2ME see the list of documents here.