Optimisation of SMART-9N for viral metagenomics
Metagenomics enables the analysis of the entire nucleic acid contents of a sample without target-specific amplification. Therefore, it presents itself as an untargeted strategy for viral surveillance, allowing the detection of both known and emerging species and variants. However, sensitivity proves challenging due to the complex microbiomes of samples and high host nucleic acid backgrounds. To overcome these setbacks, host nucleic acid depletion can be deployed, and viral RNA and DNA can be amplified.
For RNA virus detection and sequencing, SMART-9N has been utilised successfully, recovering 10 kb of the Zika virus genome in a single sequence read. SMART-9N builds on the SMART (Switching Mechanism at the 5′ end of RNA Template) concept, utilising 9N primers to randomly prime RNA for cDNA synthesis. Traditionally, SMART targets polyadenylated RNA, however, the 9N primer utilised in SMART-9N binds independently of the poly(A) tail. During reverse transcription, non-templated cytosines are added at the 3’ end of the cDNA strand. SMART-9N utilises a template-switching primer containing riboguanosines to bind to the cytosines allowing the reverse transcriptase enzyme to add the primer sequence to the 5’ end. Therefore, both ends of the cDNA contain the specific primer sequence that can be used to amplify the cDNA and enrich for RNA viruses without virus-specific primers. Furthermore, as the primers utilised are independent of the poly(A) tail of RNA, they can also anneal to and amplify DNA in a similar mechanism as described in SISPA-Seq. Hence, SMART-9N facilitates unbiased, high-resolution metagenomic sequencing for both DNA and RNA viruses.
Recently, the SMART-9N protocol has been optimised and updated to ensure that the methodology utilised produces the best results for viral metagenomics. The following optimisations have been implemented:
Host Depletion and Extraction:
-
The original protocol involves a spin-column extraction followed by DNase treatment and a clean-up column.
-
The optimised protocol performs DNase treatment first, followed by a magnetic bead extraction.
-
This facilitates the removal of extracellular DNA present in the sample and allowing DNA viruses to be processed through the extraction and hence, amplified in the SMART-9N reaction.
Primer Concentration:
-
Previously, the original protocol utilised a primer concentration of 2 µM for annealing and cDNA synthesis, followed by 20 µM of PCR primer.
-
The optimised protocol utilises a primer concentration of 12 µM for annealing and cDNA synthesis, followed by 10 µM of PCR primer.
-
The amended concentrations produced a greater yield and genome coverage in comparison to the original concentrations.
Rapid SMART-9N:
-
The original protocol utilised the ONT RLB barcoding primers (SQK-RPB114.24) to perform amplification and barcoding in a single reaction.
-
However, the ONT RLB barcoding primer fails to sufficiently amplify the cDNA, in comparison to an unmodified PCR primer that produces a ten-fold greater yield.
-
Therefore, the optimised protocol utilises the unmodified PCR primer and requires separate library preparation to barcode and adapt the samples.
The updated protocol is published on protocols.io: doi.org/10.17504/protocols.io.rm7vz9xn5gx1/v1
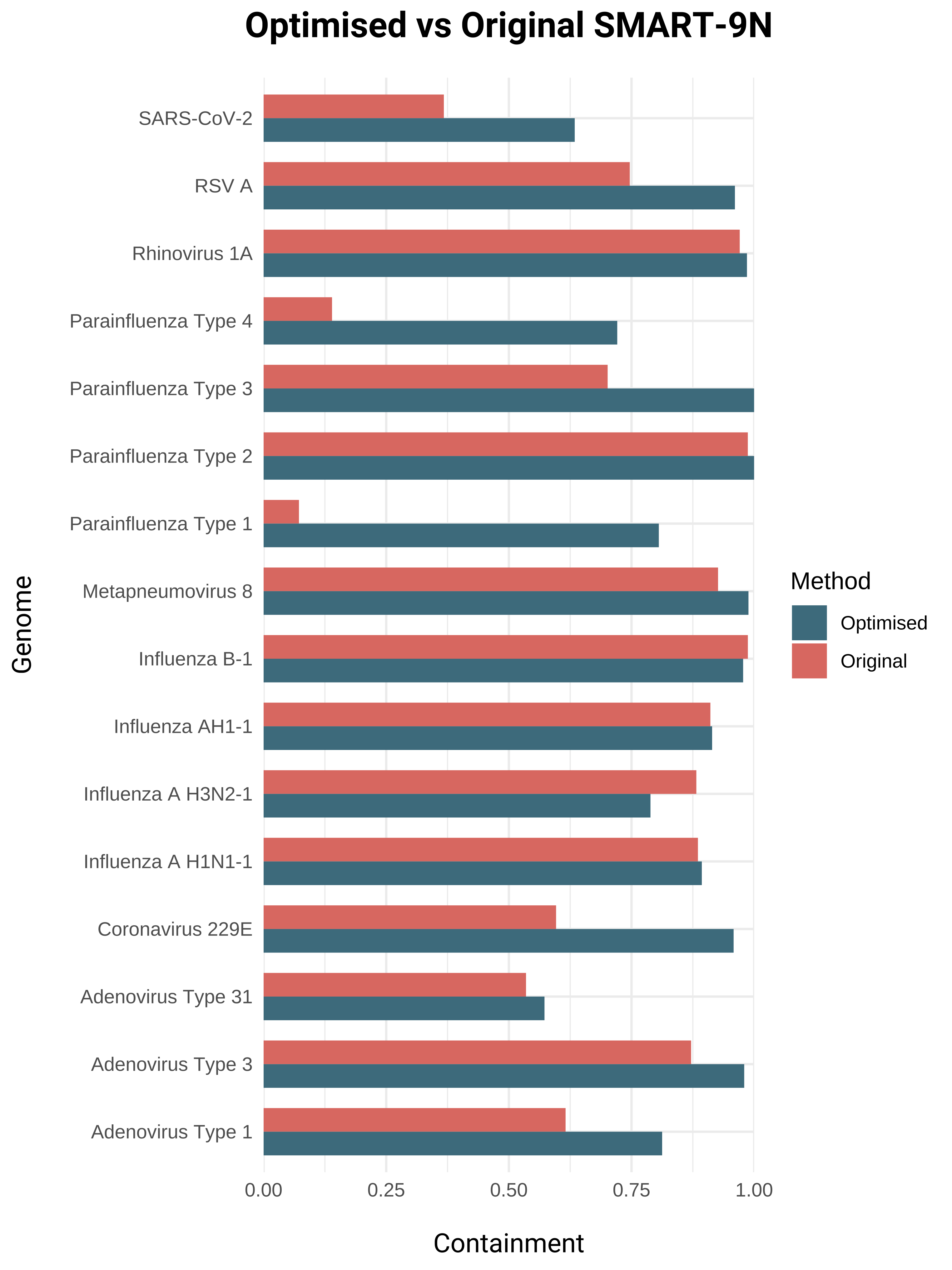
The ZeptoMetrix® NATtrol™ Respiratory Panel 2.1 (RP2.1), containing 18 viral and 4 bacterial taxa, was utilised to compare the original SMART-9N protocol to the optimised version, subsampled to 10 gbp. K-mer containment, a useful metric to assess the completeness and accuracy of assembled genomes, was used to compare the methods directly. Overall, the optimised protocol proves to recover a greater containment of the respiratory genomes present in the control.