A collection of resources for whole genome sequencing and analysis of Measles virus (MeV)
Resources and documents
Background
Measles virus genomic epidemiology
A primer on measles virus genomic epidemiology and why we do it
Primer scheme
artic-measles/400/v1.0.0 Primer Scheme
The ARTIC-network primer scheme for sequencing MEV
Wet-lab Protocol
ARTIC-network MEV wet-lab protocol
The ARTIC-network wet-lab protocol for sequencing MEV using the artic-measles/400/v1.0.0 primer scheme
User-interface pipelines using Epi2me
EPI2ME Installation
A guide to help you install and set up EPI2ME from the EPI2ME team
amplicon-nf: Running the pipeline in EPI2ME
Using EPI2ME for running the ARTIC amplicon-nf pipeline without the commandline
amplicon-nf: Running the pipeline in EPI2ME for Illumina Data
Using EPI2ME for running the ARTIC amplicon-nf pipeline without the commandline
amplicon-nf: Running the pipeline in EPI2ME for Oxford Nanopore Data
Using EPI2ME for running the ARTIC amplicon-nf pipeline without the commandline
amplicon-nf: Viewing outputs of the pipeline in EPI2ME
Viewing outputs of the amplicon-nf pipeline in EPI2ME
Command line interface pipeline SOPs
Fieldbioinformatics: viral amplicon sequencing bioinformatics SOP
Nanopore | bioinformatics
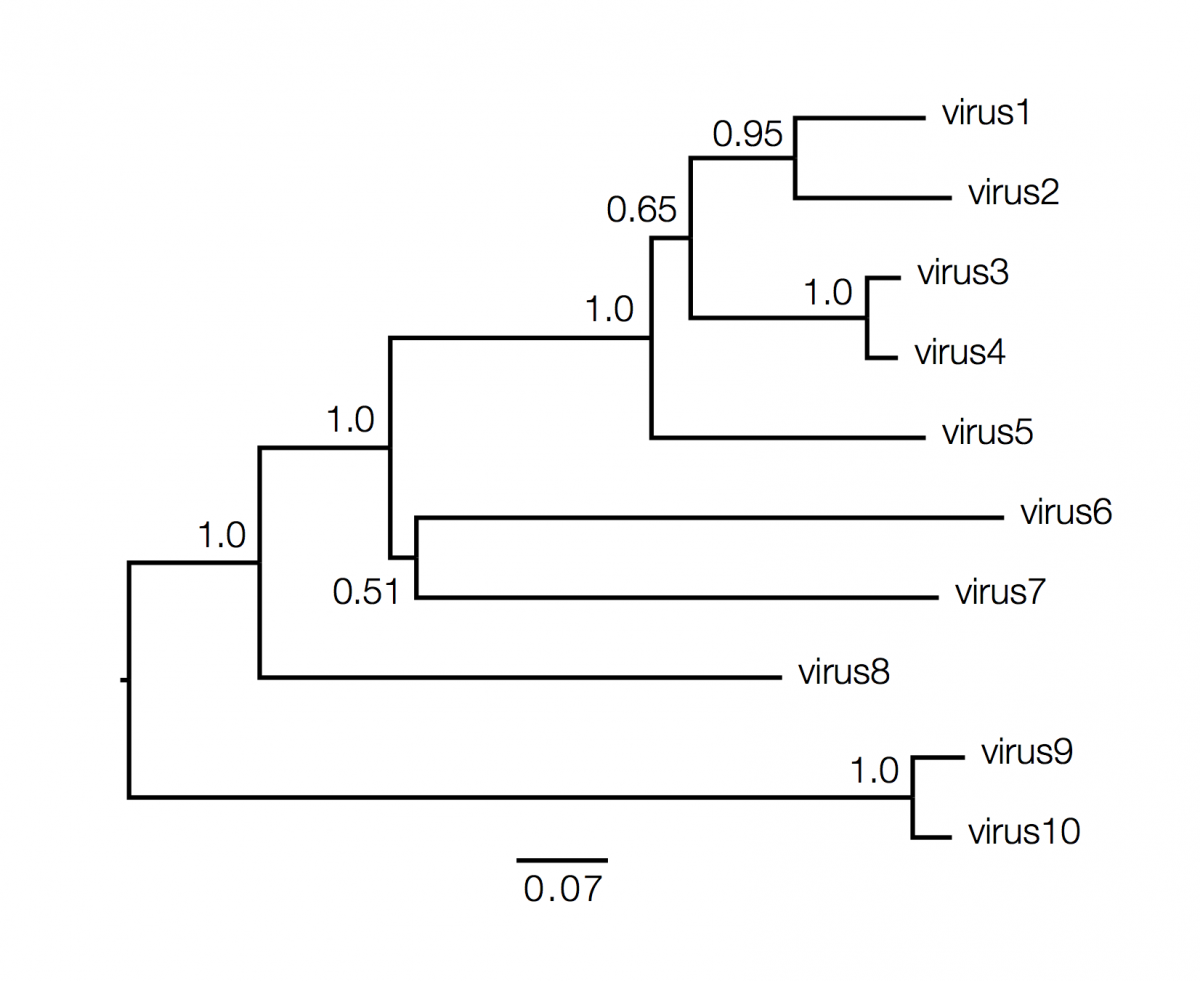
Phylogenetics and Interpretation
MeV Phylogenetics
Resources and protocols for MeV alignment and phylogenetics